Growing portfolio of recently collected and fully characterized PDXs to advance personalized medicine approaches
Choose from over 300 models originating within the last 7 years for a more accurate reflection of human cancer biology compared to older or less relevant models. Our growing portfolio of models range multiple histotypes and biomarkers.
Cancer types include:
- Brain
- Breast
- Cholangio/Bile duct
- Colorectal
- Gastric
- Head and neck
- Lung
- Mesothelioma
- Neuroendocrine
- Ovarian
- Pancreatic
- Renal
- Skin
- Sarcoma
- Uveal
Biomarkers include, but not limited to:
- Claudin 6
- TROP2
- EGFR
- HER2
- KRAS
- CCNE1
- BRCA1/2
- MUC16
-
Disease-relevant PDXs representing multiple histotypes
-
Extensive molecular and gene expression characterization in addition to treatment history
-
Tumor models fresh from the clinic at low passage
Maximize the insights gained from PDX Models
Our optimized PDX engraftment procedures and modern technologies, such as multi-omics, digital spatial analysis and biodistribution, provide comprehensive insights into their molecular landscape. To utilize the unique genomic features of PDX models we offer a broad range of in vivo and ex vivo analysis capabilities. Our solutions include a variety of therapeutic approaches including adoptive cell therapies and combination therapies with radiation and radiopharmaceuticals to support your oncology research program.
Oncology experience you can count on
With more than 10,000 studies completed, we have amassed extensive experience and deep insights to inform end-to-end discovery oncology research. Our team can help:
- Explore the effects of drugs and biologics using both human and syngeneic tumor models.
- Empower you to make more informed decisions at every step of the development process.
- Support you with testing throughout the entire development journey, including bioanalysis, pharmacology, toxicology, companion diagnostics, central labs, and more.
- Gain efficiencies as we connect insights effectively from one stage to the next.
PDX FAQs
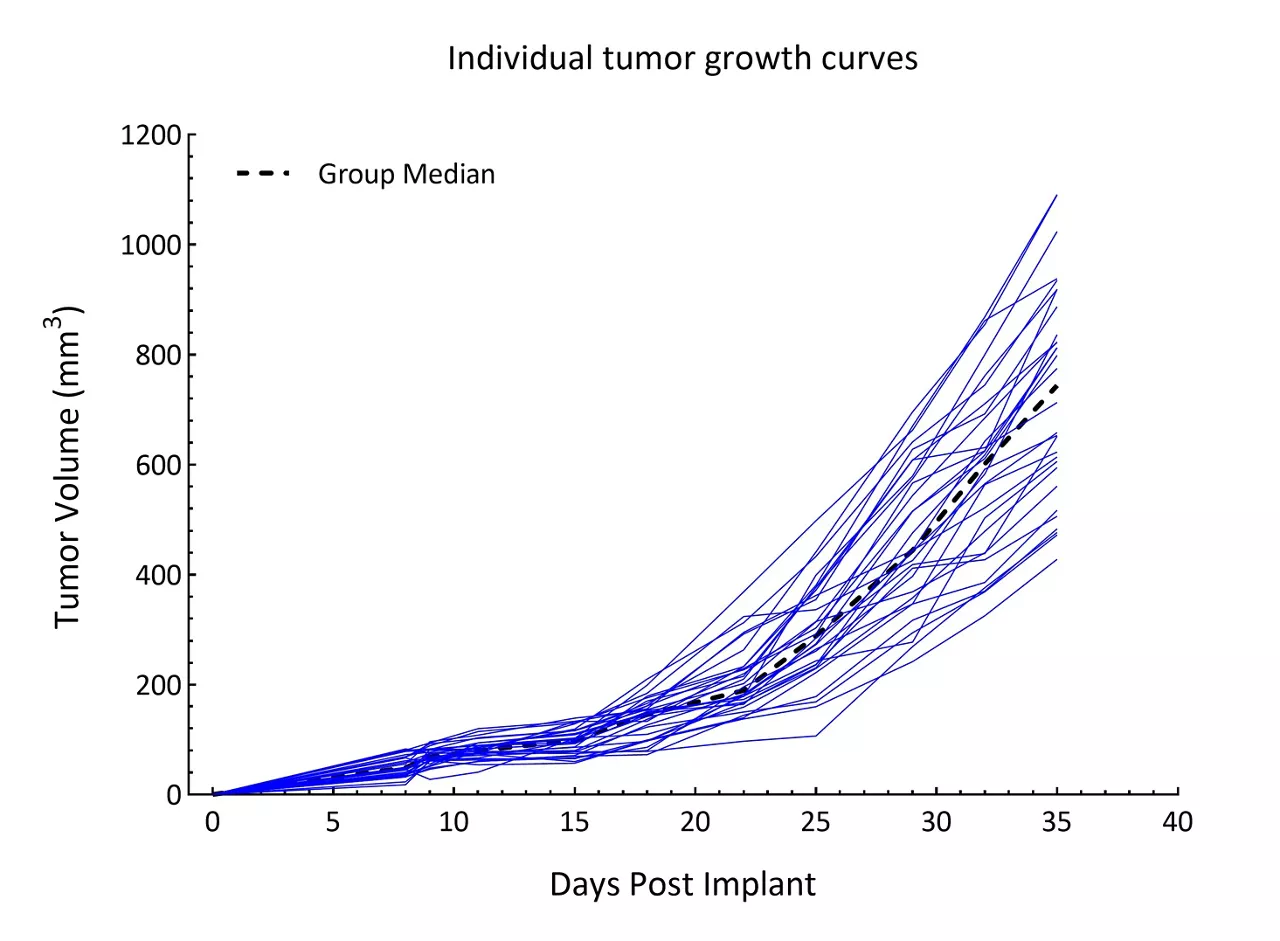
Patient-derived xenograft (PDX) models are cutting-edge tools in oncology research, derived from transplanting patient tumor tissues directly into immunocompromised mice. These models preserve the genetic, histological, molecular and spatial characteristics of the original human tumors including intratumor heterogeneity, offering a highly relevant platform for preclinical drug testing.
- Clinical relevance: PDX models closely mimic human tumors, including tumor heterogeneity, providing a superior model compared to traditional cell lines or genetically engineered models
- Predictive power: PDX models predict patient responses to therapies more accurately than other models, enabling researchers to prioritize drug candidates with higher potential for clinical success
- Personalized medicine: Each PDX model is derived from a specific patient's tumor, allowing for testing of therapies tailored to individual genetic and molecular profiles
- Study of resistance mechanisms: PDX models facilitate the study of drug resistance mechanisms by enabling longitudinal studies of tumor progression and response to treatment over time
- Long-term stability: PDX models can be passaged through multiple generations while maintaining the genetic and phenotypic stability of the original tumor, supporting longitudinal studies and reproducible results
Standard use includes, but is not limited to:
- Preclinical evaluation of novel therapies (including cell therapy)
- Validating novel drug combinations
- Screening drug-sensitive patients (personalized medicine)
- Exploring drug response/resistance mechanisms
- Biomarker evaluation


?fmt=webp)